Recent publications from the CMU biophysics group – not a complete list. A more comprehensive overview of publications is provided on the group pages of the individual investigators – and on Google Scholar, the National Library of Medicine (NCBI), ORCID, or Publons (aka ResearcherID).

Fundamental principles in bacterial physiology—history, recent progress, and the future with focus on cell size control: a review
Suckjoon Jun, Fangwei Si, Rami Pugatch, & Matthew Scott
This 64-page article reviews more than 700 references in the history of bacterial physiology since the beginning of the 20th century, providing a comprehensive overview of major ideas and approaches for anyone interested in the fundamental problems in bacterial physiology.
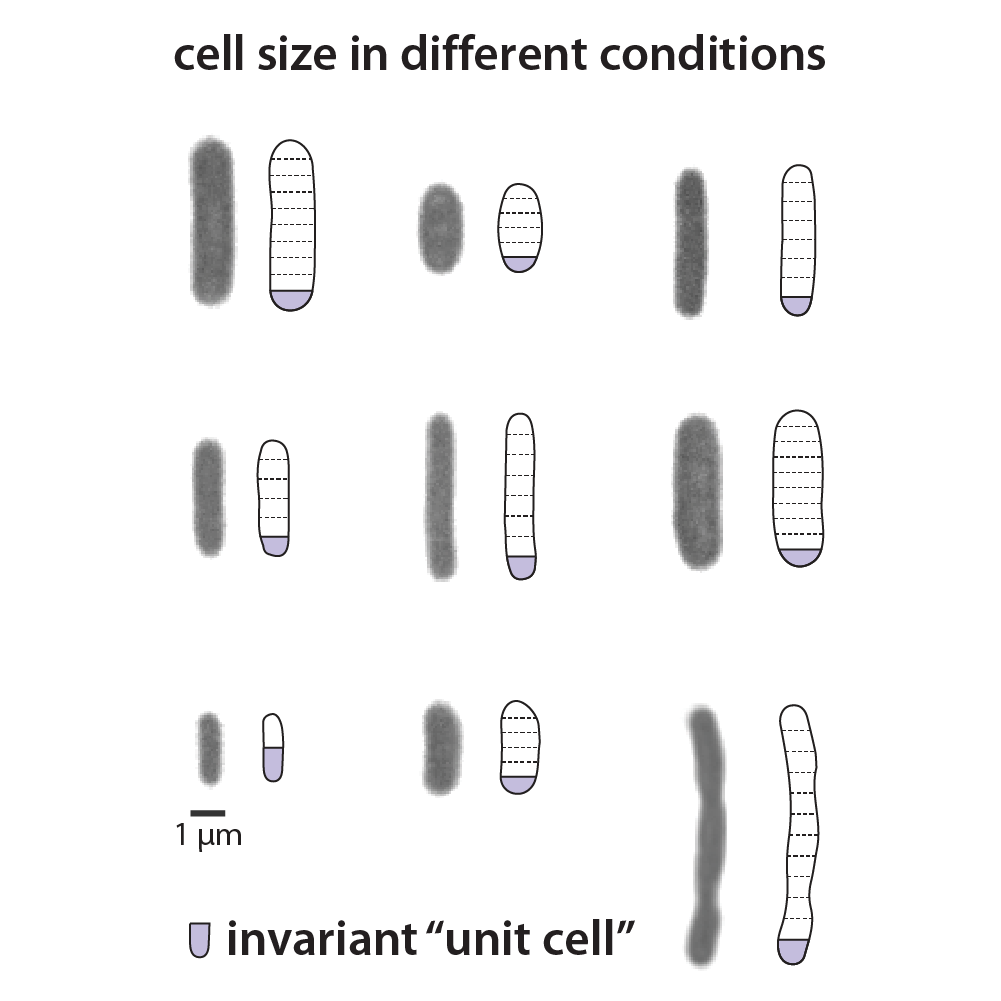
Invariance of initiation mass and predictability of cell size in Escherichia coli
Fangwei Si, Dongyang Li, Sarah E. Cox, John T. Sauls, Omid Azizi, Cindy Sou, Amy B. Schwartz, Michael J. Erickstad, Yonggyn Jun, Xintian Li, & Suckjoon Jun
In this paper, we hypothesized that the E. coli cell is composed of conceptual “unit cells.” By extensively perturbing the growth physiology in steady-state conditions using genetic and pharmacological methods, we discovered the invariant initiation mass as the basic unit of the cell size. This finding unraveled how cells coordinate their biomass and DNA content by imposing a simple control on replication initiation.
See also the associated commentary in Current Biology, as well as the news Coverage by phys.org and ScienceDaily.
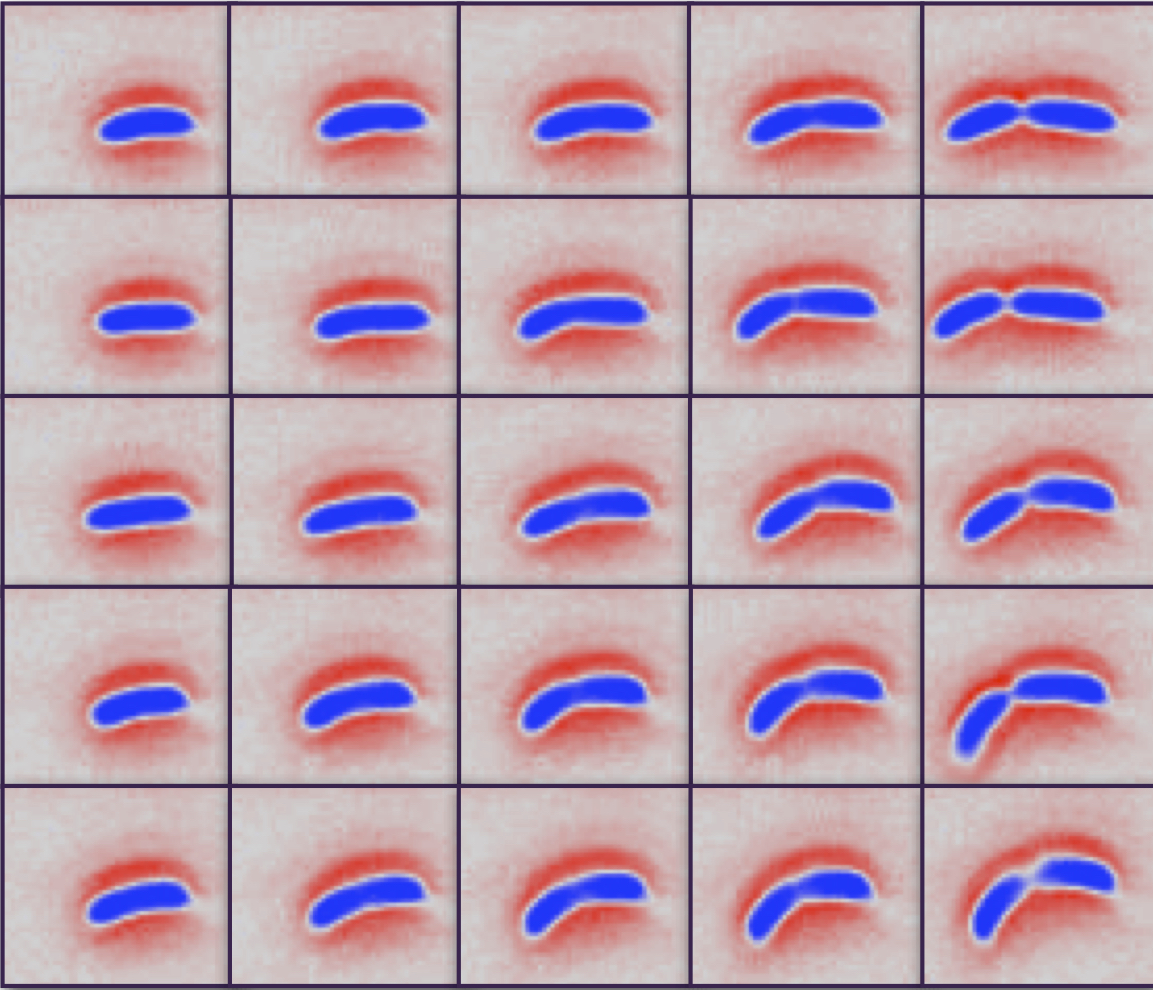
Biphasic growth dynamics control cell division in Caulobacter crescentus
S. Banerjee, K. Lo, M.K. Daddysman, A. Selewa, T. Kuntz, A.R. Dinner and N.F. Scherer
Cell size is specific to each species and impacts cell function. Here, we investigate size control and the cell cycle dependence of bacterial growth using multigenerational cell growth and shape data for single C. crescentus cells. Our analysis reveals a biphasic mode of growth: a relative timer phase before cell wall constriction, followed by a pure adder phase during constriction. We present a mathematical model that quantitatively explains this biphasic model for cell size control.
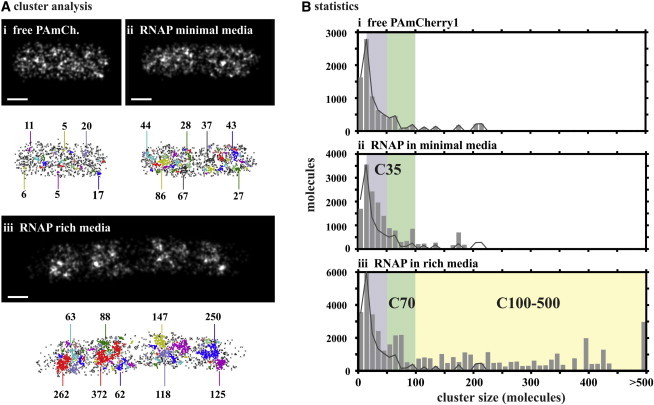
Multiscale spatial organization of RNA polymerase in Escherichia coli
U. Endesfelder, K. Finan, S. Holden, P. Cook, A. Kapanidis, and M. Heilemann
We use photoactivated localization microscopy to localize individual molecules of RNA polymerase (RNAP) in Escherichia coli. In cells growing rapidly in nutrient-rich media, we find that RNAP is organized in 2–8 bands. The band number scaled directly with cell size (and so with the chromosome number), and bands often contained clusters of >70 tightly packed RNAPs and clusters of such clusters. In nutrient-poor media, RNAPs were located in only 1–2 bands; within these bands, a disproportionate number of RNAPs were found in clusters containing ∼20–50 RNAPs.
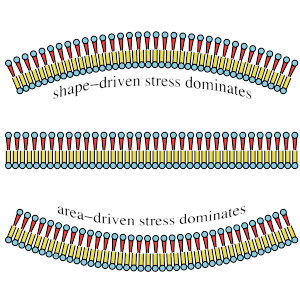
Spontaneous curvature, differential stress, and bending modulus of asymmetric lipid membranes
A. Hossein and M. Deserno
The two leaflets of a lipid bilayer can differ in composition or in stress, but these two degrees of asymmetry are coupled. They can affect curvature and even rigidity of a membrane. This paper discusses the connection, and how it is affected by a lipid species, such as cholesterol, that can flip-flop between leaflets.
See also New and Notable by Sodt and Lyman.
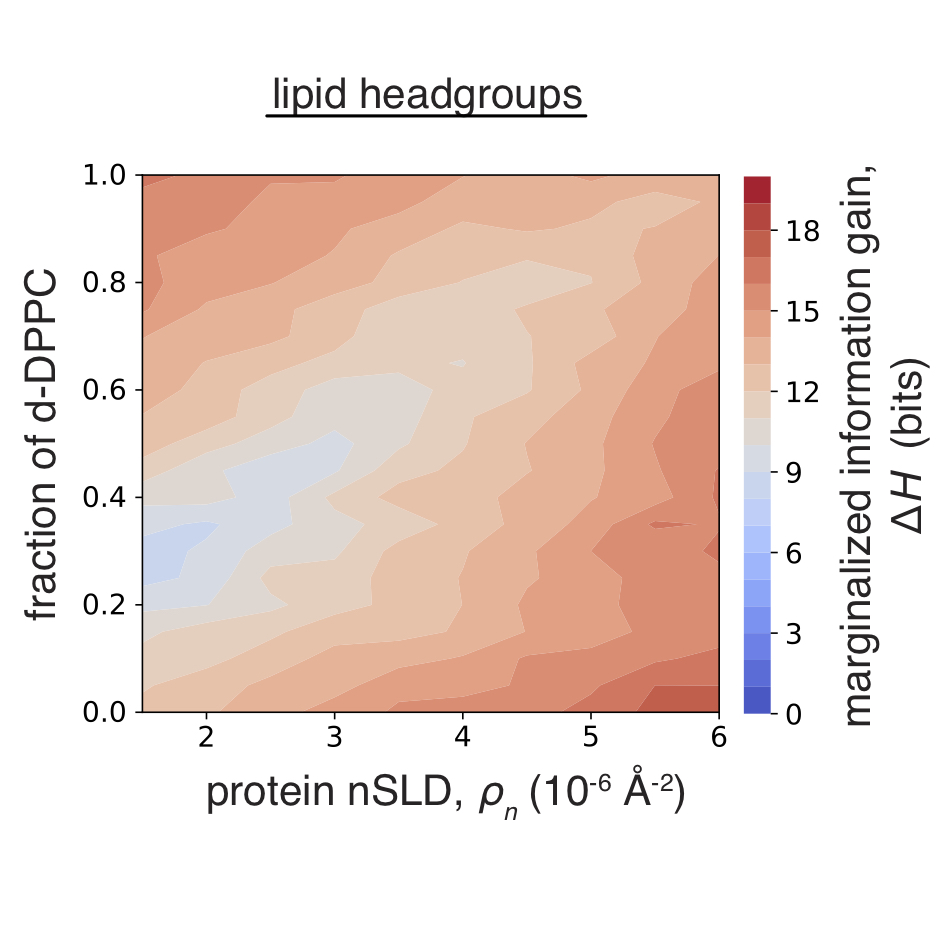
Information gain from isotopic contrast variation in neutron reflectometry on protein-membrane complex structures
F. Heinrich, P. A. Kienzle, D. P. Hoogerheide, and M. Lösche
A new information theory-based framework is applied to quantify information gain from neutron or X-ray reflectometry experiments in an in-depth investigation into the design of scattering contrast in biological and soft-matter surface architectures. To focus the experimental design on regions of interest, the marginalization of the information gain with respect to a subset of model parameters describing the structure is implemented. Surface architectures of increasing complexity from a simple model system to a protein–lipid membrane complex are simulated.
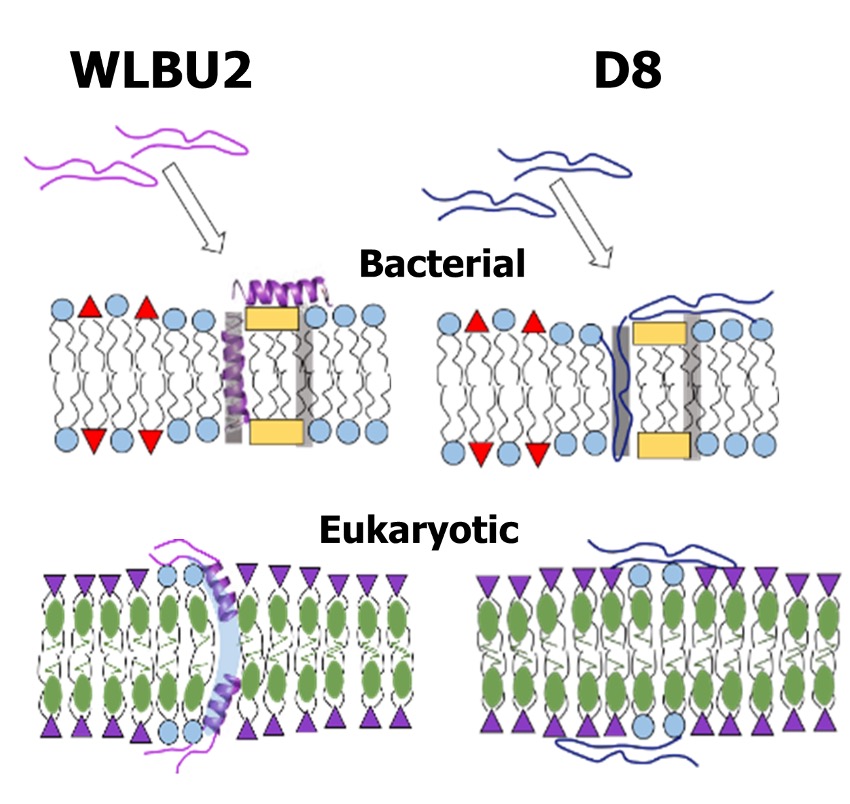
Synergistic biophysical techniques reveal structural mechanisms of engineered cationic antimicrobial peptides in lipid model membranes
F. Heinrich, A. Salyapongse, A. Kumagai, F.G. Dupuy, K. Shukla, A. Penk, D. Huster, R.K. Ernst, A. Pavlova, J.C. Gumbart, B. Deslouches, Y.P. Di, and S. Tristram-Nagle
In the quest for new antibiotics, two novel antimicrobial peptides have been rationally designed. We explore protein secondary structure, location of peptides in six lipid model membranes, changes in membrane structure and pore evidence. Our results suggest that protein secondary structure is not a critical determinant of bacteri- cidal activity, but that membrane thinning and dual location in the membrane headgroup and hydrocarbon regions is important.
Association of model neurotransmitters with lipid bilayer membranes
Receptor ion channels in the postsynaptic membrane and their neurotransmitter agonists enable fast communication between neuronal cells. Here we use thermodynamic and structural measurements to quantify the membrane-bound states of the neurotransmitter serotonin.
Mechanical Properties of Lipid Bilayers:
A Note on the Poisson Ratio
M.M. Terzi, M. Deserno, and J.F. Nagle
The Poisson ratio of elastic materials quantifies how uni- or biaxial strains induce strains in the remaining perpendicular dimension(s). Do membranes have a Poisson ratio? And given that membranes have strong material gradients along their normal, does it vary with position?

M.M. Terzi, M.F. Ergüder, and M. Deserno
Curvature is the essential geometric degree of freedom of a membrane. But at next order the lipid’s orientation adds two more degrees of freedom: tilt and twist. Here we discuss how to construct a theory for all of them that is consistent at the quadratic level and compare it with extensive atomistic simulations.
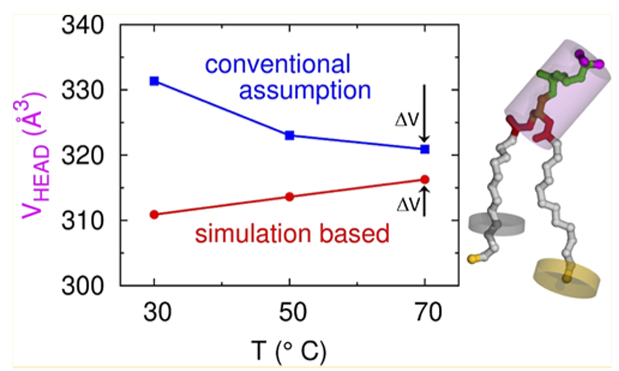
Revisiting Volumes of Lipid Components in Bilayers
J.F. Nagle, R.M. Venable, E. Maroclo-Kemmerling, S. Tristram-Nagle, P.E. Harper, and R.W. Pastor
In addition to obtaining the highly precise volumes of lipids in lipid bilayers, it has been desirable to obtain the volumes of parts of each lipid. We propose a new method to refine the determination of component volumes, which reveals that head group free volumes are relatively independent of thermodynamic phase, whereas both the methylene and methyl free volumes increase dramatically when bilayers transition from gel to fluid.
Elastic Behavior of Model Membranes with Antimicrobial Peptides Depends on Lipid Specificity and D-enantiomers
A. Kumagai, F.G. Dupuy, Z. Arsov, Y. Elhady, D. Moody, R.K. Ernst, B. Deslouches, R.C. Montelaro, Y.P. Di, S. Tristram-Nagle
In an effort to provide new treatments for the global crisis of bacterial resistance to current antibiotics, we have used a rational approach to design several new antimicrobial peptides. We show a good correlation between in vitro bacterial killing efficiency and both bending and chain order behavior in bacterial lipid membrane mimics. These results suggest that elasticity and chain order behavior can be used to predict mechanisms of bactericidal action.
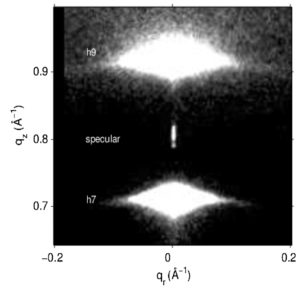
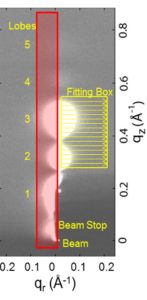
Structure of gel phase DPPC determined by X-ray diffraction
J.F. Nagle, P. Cogneta, F.G. Dupuy, S. Tristram-Nagle
High resolution low angle x-ray data are reported for the gel phase of DPPC lipid bilayers, extending the previous range of wavevectors, and employing a new technique to obtain more accurate intensities and form factors for the highest orders of diffraction.
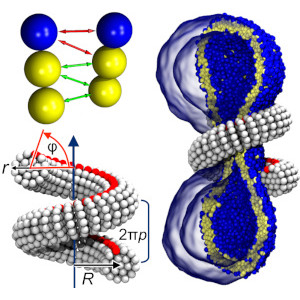
M. Pannuzzo, Z.A. McDargh, and M. Deserno
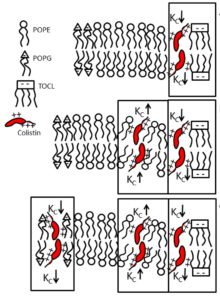
Selective Interaction of Colistin with Lipid Model Membranes
F.G. Dupuy, I. Pagano, K. Andenoro, M.F. Peralta, Y. Elhady, F. Heinrich, S. Tristram-Nagle.
To provide molecular details about colistin’s ability to kill Gram-negative, but not Gram-positive bacteria, we investigated the interaction between colistin and lipid mixtures mimicking the cytoplasmic membrane of G(+) and G(–) bacteria as well as eukaryotic cells. Two different models of the G(–) outer membrane were assayed: lipid A with two deoxy-manno-octulosonyl sugar residues, and Escherichia coli lipopolysaccharide mixed with dilaurylphosphatidylglycerol. Our results suggest that domain formation is responsible for the erratic response of G(–) inner membranes to colistin and for its deeper penetration, which could increase membrane permeability.
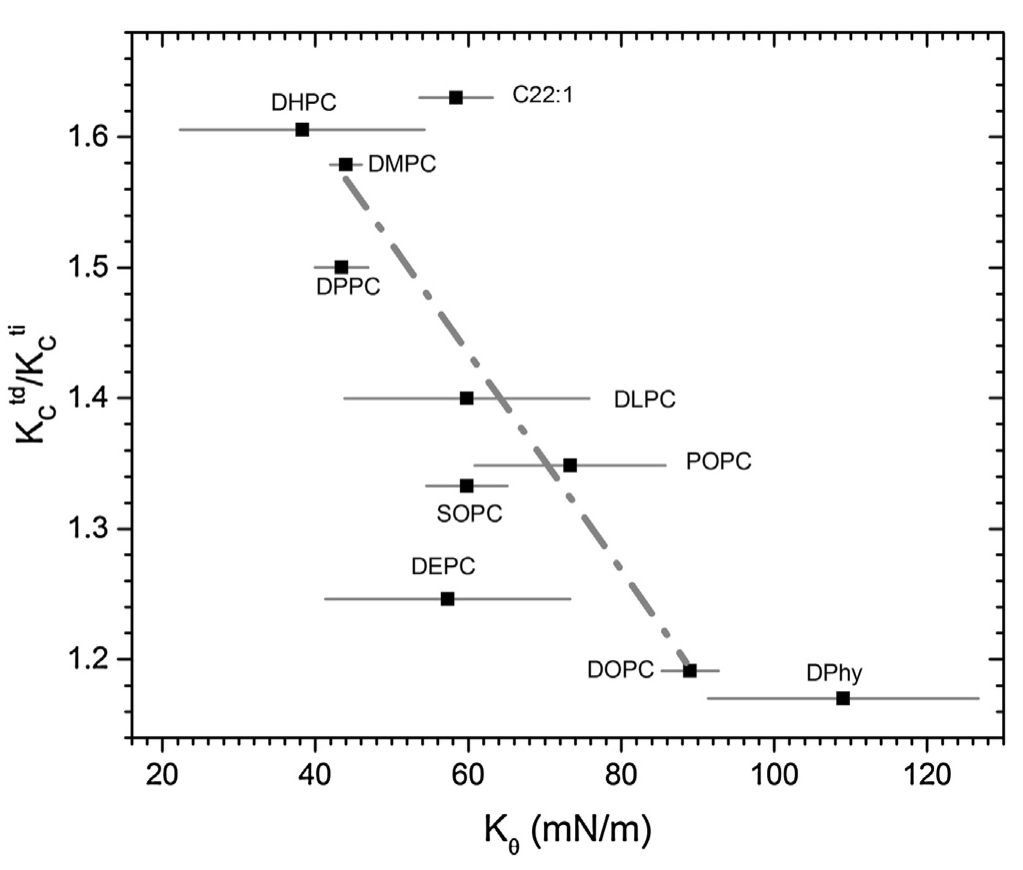
Experimentally determined tilt and bending moduli of single-component lipid bilayers
J.F. Nagle
The paper presents the first experimental determination of the tilt modulus for many single component lipid bilayers.
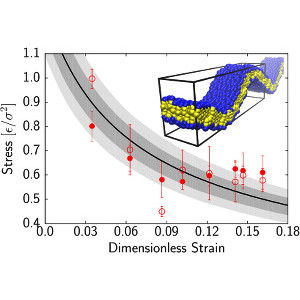
P. Diggins IV, Z.A. McDargh, and M. Deserno
The collective behavior of cells in epithelial tissues is dependent on their mechanical properties. Here we show that epithelial tissues can transition from a solid-like jammed state to a fluid-like unjammed state to accelerate the rate of wound healing and tissue repair.
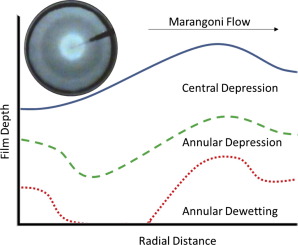
Flow regime transitions and effects on solute transport in surfactant-driven Marangoni flows
Surfactant-driven Marangoni flow on liquid films impacts the transport of soluble species entrained in the flow. Understanding the flow behavior is important for applications in drug delivery, coatings, and oil spill remediation. Here we develop a novel technique to quantitatively measure the Marangoni flow and map the conditions at which key flow regime transitions occur.
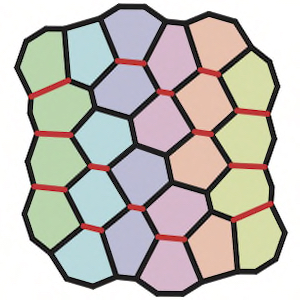
M.F. Staddon, K.E. Cavanaugh, E.M. Munro, M.L. Gardel, and S. Banerjee
During development, epithelial tissues form complex structures like organs through precise spatiotemporal coordination of cell shape changes. But the forces underlying these irreversible shape changes remain elusive. Here, we demonstrate that pulsatile contraction forces ensure robust cell shape changes via a mechanical ratchet.
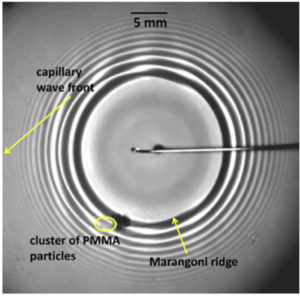
Surfactant-induced Marangoni transport of lipids and therapeutics within the lung
A.Z. Stetten, S.V. Iasella, T.E. Corcoran, S. Garoff, T.M. Przybycien, R.D. Tilton
Aerosolizing Lipid Dispersions Enables Antibiotic Transport Across Mimics of the Lung Airway Surface Even in the Presence of Pre-existing Lipid Monolayers
S.V. Iasella, A.Z. Stetten, T.E. Corcoran, S. Garoff, T.M. Przybycien, R.D. Tilton
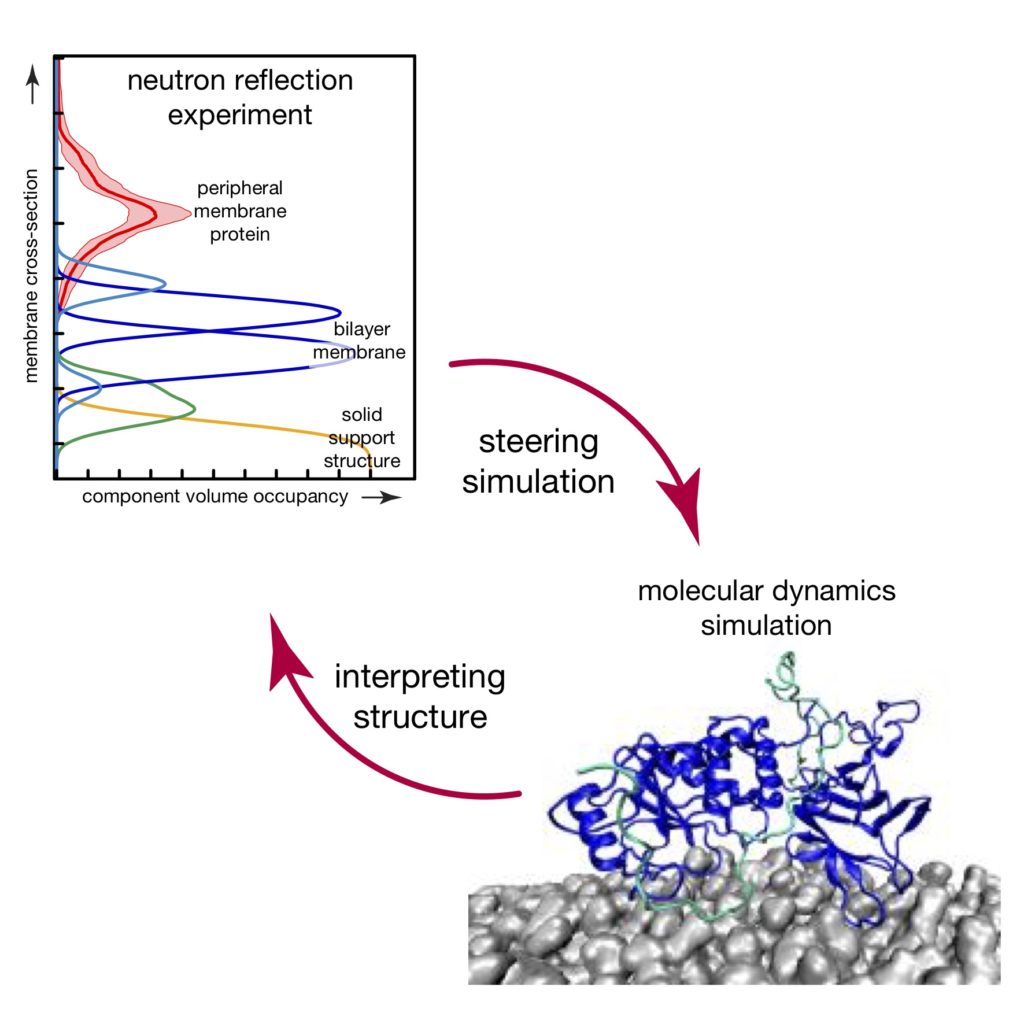
B.W. Treece, F. Heinrich, A. Ramanathan, and M. Lösche
We present a novel method to incorporate structural results from surface-sensitive scattering, such as X-ray or neutron reflectometry, into molecular dynamics simulations. This method is particularly valuable for studies of proteins at interfaces that contain disordered regions since the conformation of such regions is difficult to judge from the analysis of one-dimensional experimental profiles and may take prohibitively long to equilibrate in simulations.

Mechanistic origin of cell-size control and homeostasis in bacteria
Fangwei Si, Guillaume Le Treut, John T. Sauls, Stephen Vadia, Petra Anne Levin, & Suckjoon Jun
In this paper, we proposed the hypothesis that bacterial cell division requires the accumulation of division proteins to a fixed threshold number. By combining single-cell microfluidics, programmed genetic modulation, and automated image analysis, we confirmed this hypothesis in both E. coli and B. subtilis cells. This latter result explains mechanistically why a wide range of bacterial species follow the “adder” principle of cell-size homeostasis.
See also the associated video abstract, as well as the news coverage in phys.org.
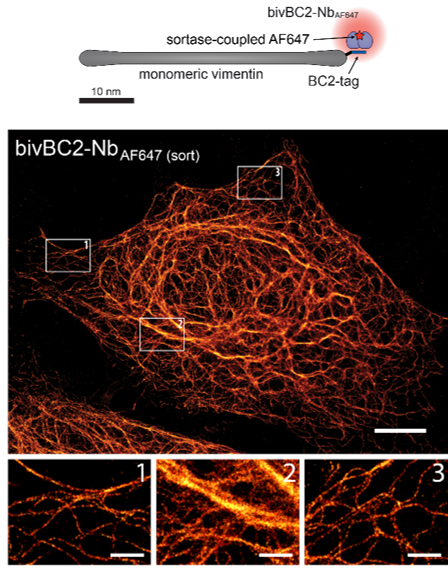
D. Virant, B. Traenkle, J. Maier, P. D. Kaiser, M. Bodenhöfer, C. Schmees, I. Vojnovic, B. Pisak-Lukáts, U. Endesfelder, U. Rothbauer
Dense fluorophore labeling without compromising the biological target is crucial for genuine super-resolution microscopy. Here we introduce a broadly applicable labeling strategy for fixed and living cells utilizing a short peptide tag-specific nanobody (BC2-tag/bivBC2-Nb). BC2-tagging of ectopically introduced or endogenous proteins does not interfere with the examined structures and bivBC2-Nb staining results in a close-grained fluorophore labeling with minimal linkage errors. This allowed us to perform high-quality dSTORM imaging of various targets in mammalian and yeast cells.
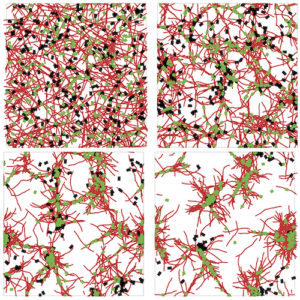
A versatile framework for simulating the dynamic mechanical structure of cytoskeletal networks
S.L. Freedman, S. Banerjee, G.M. Hocky, and A.R. Dinner
Computer simulations can aid in understanding how collective materials properties emerge from interactions between simple constituents. Here, we introduce a coarse-grained model that enables simulation of networks of actin filaments, myosin motors, and cross-linking proteins at biologically relevant time and length scales.
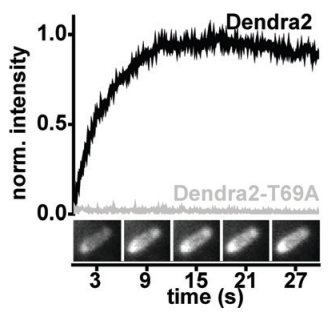
A general mechanism of photoconversion of green‐to‐red fluorescent proteins based on blue and infrared light reduces phototoxicity in live‐cell single‐molecule imaging
B. Turkowyd, A. Balinovic, D. Virant, H. G. Gölz Carnero, F. Caldana, M. Endesfelder, D. Bourgeois, U. Endesfelder
Photoconversion of fluorescent proteins by blue and complementary near‐infrared light, termed primed conversion (PC), is a mechanism recently discovered for Dendra2. We demonstrate that controlling the conformation of arginine at residue 66 by threonine at residue 69 of fluorescent proteins from Anthozoan families (Dendra2, mMaple, Eos, mKikGR, pcDronpa protein families) represents a general route to facilitate PC. Mutations of alanine 159 or serine 173, which are known to influence chromophore flexibility and allow for reversible photoswitching, prevent PC. In addition, we report enhanced photoconversion for pcDronpa variants with asparagine 116. We demonstrate live‐cell single‐molecule imaging with reduced phototoxicity using PC.
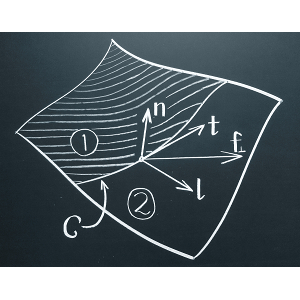
M. Deserno
Differential geometry is an enormously powerful tool for describing membranes, which at sufficidently large scales are just curved surfaces embedded in three-dimensional space. This review gives a detailed and pedagogical overview of the essential mathematics, and then applies it to the very powerful notion of membrane stresses.
