Problems under study at the Biological Physics Group at Carnegie Mellon include: quantifying single cells’ behaviors and sub-cellular structure and dynamics, physics of biological membranes, physical principles of membrane self-assembly, membrane-protein interactions, the molecular basis of cell signaling, mechanics of the cytoskeleton, cell motility and tissue morphogenesis.
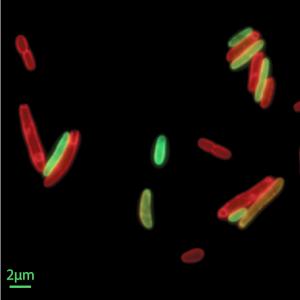
Single-Cell Biophysics
A major challenge in biophysics is to decode the molecular processes underpinning physical behaviors at the cellular scale: how cells grow, how they divide, how they change their shapes to move, and how cells respond to their environment. To develop a mechanistic understanding of single-cell behavior we develop theory, design new experiments and computational methods for single-cell analyses. Topics of interest include studying the spatial organization and dynamics of sub-cellular structures, and how these control cellular processes. To this end, the Si lab develops and adapts single-cell techniques, such as microfluidics, to obtain high-quality data that can help reveal quantitative relationships between the complicated cell fitness and form. On the theory side, Banerjee lab develops computational models to relate molecular-scale dynamics with cellular-scale physical behavior, including cell growth, motility and replication cycle.
Membrane Biophysics
Lipid membranes form the boundaries of all living cells, and many internal organelles in nucleated cells. They are molecularly thin fluid elastic films with amazing material properties that underly their biological function, many of which pose unanswered biophysical and biological questions to this day. Why are there so many different types of lipids? How do proteins insert into, bind onto, and fold inside membranes? How do cell membranes maintain their asymmetry? How do elastic properties emerge from their self-assembled components? How are stresses transmitted along membranes? – Our team has many years of experience in studying lipid membranes using experiment, theory, and computation. Using diffuse X-ray scattering, neutron reflectometry, densitometry, surface plasmon resonance, molecular dynamics simulation, systematic coarse-graining, continuum theory, differential geometry, and statistical field theory we apply a wide set of tools to learn more about the thin films that separate life from death.
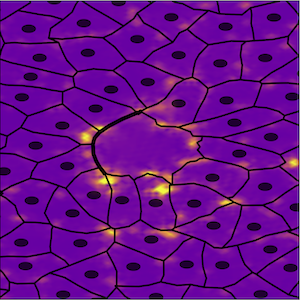
Physics of Collective Behavior
Living systems exhibit complex self-organized behavior that results from coordinated interactions between its individual components. The emergence of complex collective behaviors occurs in biological systems over a wide range of scales – from lipids that organize cell membranes to protein-based polymers and protein motors that assemble the cell cortex and groups of cells that form complex tissues. A fundamental problem we seek to understand is how collective physical properties emerge in living systems from dynamic interactions between individual parts. To this end we develop multi-scale theory and computational models of collective dynamical systems, leveraging progress in soft matter physics, statistical mechanics and experimental biology. Current research in our group involves understanding the self-organization and mechanics of lipid membranes, the dynamics and structure of the cell cytoskeleton, tissue mechanics and collective cell dynamics.
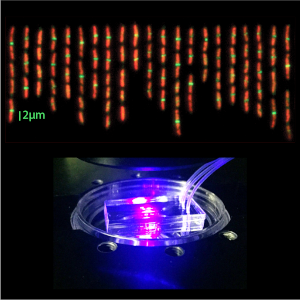
Biophysical Tools Development
Our projects offer incentives, test cases and applications for the development of a suite of new biophysical tools as a practical approach to biophysical research. These include single-cell microfluidics and microscopy, X-ray and neutron scattering, statistical physics, non-equilibrium dynamics, differential geometry, field theory, machine learning and molecular dynamics simulations.
Our recent developments include:
- Single-cell microfluidic tool that quantifies the binding affinity of peripheral membrane proteins in individual live cells
- Membrane perturbation methods to manipulate basic properties of the membranes in individual live cells, such as membrane’s protein-to-lipid ratio and the capacity of membrane biogenesis.
- Data modeling in surface-sensitive scattering: A steering potential for MD simulations based upon neutron reflectometry data
- Simulation packages for modelling cell and tissue dynamics: active filament network simulations, the Active Vertex Model
- Analysis of scattering experiments using information theory: Method development and application to protein-membrane complexes
