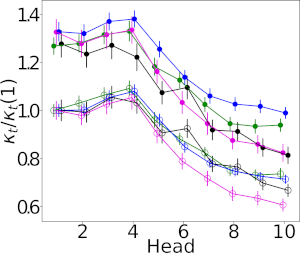
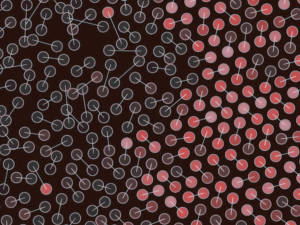
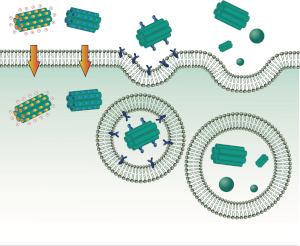
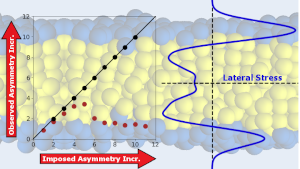
Welcome…
…to the web pages of the group of Theoretical and Computational Biological Physics, established at the Department of Physics at Carnegie Mellon in Fall 2007!
from left to right: Nick, Mert, Markus, Martina, Muhammed, Nishchay, Amirali
In our research we use theoretical and computational tools to understand the behavior of a variety of biological systems. We are particularly interested in biological membranes and proteins. Our main tools are coarse grained molecular simulations, continuum theory and statistical physics.
Feel free to browse our pages and have a look at the scientific things we’re doing here!